目录
- 前言
- 一、项目背景
- 行业痛点
- 市场需求
- 二、项目概述
- 三、项目过程
- 挑战
- 少样本学习(Few-shot learning)
- 基线模型选择
- 模型改进
- (一)baseline性能
- (二)baseline+ DETR head
- (三)baseline+ RepC3K2
- 快速实现的 CSP 瓶颈模块(Faster Implementation of CSP Bottleneck)
- (四)baseline+ RepC3K2 + SimSPPF
- 简化版空间金字塔池化模块(Simplified SPP
- (五)baseline+ RepC3K2 + SimSPPF + LK-C2PSA
- 位置敏感注意力模块(Position - Sensitive Attention block)
- (六)baseline+ RepC3K2 + SimSPPF + ~~LK-C2PSA~~ TriAttentionPSA
- 基于三元组注意力(Triplet Attention)的位置敏感注意力模块
- (七)baseline+ RepC3K2 + SimSPPF + ~~LK-C2PSA~~ TriAttentionPSA+ newLabels
- 四、应用场景与价值
- 应用场景
- 总结
前言
在当今社会,中药材作为我国传统医学的瑰宝,其市场需求与日俱增。然而,中药材检测领域却面临着诸多痛点,如专业人才稀缺、质量监管难度大等。同时,随着市场规模的扩大和政策对质量追溯体系的要求,传统的检测手段已难以满足行业发展的需求。在此背景下,我们团队着手研发了“灵识本草”—— 一套融合深度学习技术的中药材高精度检测系统。本项目旨在通过先进的技术手段,提升中药材检测的精度与效率,推动中药产业的数字化升级,为中药材科研、教育、生产与加工等领域提供有力支持,助力中医药行业的现代化发展。
项目致谢:感谢团队子阳、鸿辉、嘉祥、妙君、杨琼的积极参与
一、项目背景
行业痛点
-
专业人才稀缺 :专业药材识别人才占比不足 10%(引用《2023-2024
年中国中药材行业大数据及商业趋势研究报告》),传统鉴别方法效率低、主观性强、易受炮制加工干扰。 -
质量监管难度大 :中药材质量参差不齐,传统检测手段难以满足大规模、高精度的质量监管需求。
市场需求
- 市场规模增长 :2025 年中药材市场规模预计达 5000 亿元(引用《2025-2030
年中药材产业深度调研及未来发展现状趋势预测报告》),老龄化催生慢性病管理需求,心脑血管、消化系统等中药优势领域持续扩容。 - 质量追溯需求
:《国务院办公厅关于提升中药质量促进中医药产业高质量发展的意见》要求数智技术、标准化推动全产业链升级,构建覆盖种植、加工、流通的质量追溯体系。
二、项目概述
本项目开发基于 YOLOv11 的中药材高精度智能检测系统,集成高效特征(如三重注意力机制)提取与多尺度融合(如自上而下、自下而上的融合策略)模块,实现复杂场景(堆叠 / 光照干扰)下的高精度(mAP≈86.4%)和快速(CPU 下 15fps,GPU 下 70+fps)检测,推动中药产业数字化升级。
三、项目过程
挑战
- 检测类别的多样性 :50 + 类别。
- 类别数量不均衡 :存在长尾分布,多数 label 的实例数量超过 200
- 个样本,但存在少数样本数极少,不足 10 个。
- 检测的复杂性 :目标重叠、多样背景。
少样本学习(Few-shot learning)
- 动机 :存在标签的样本数量非常稀少,例如多数 label 的实例数量超过 200 个样本,但是存在少数的样本数极少,不足 10 个。
- 优化策略 :模型结构优化学习(共 4 种策略)、数据增强策略。
基线模型选择
-
选择 YOLOv11 的原因 :YOLOv12 也是 SOTA,但考虑到测试环境下无 GPU,而 YOLOv12依赖耗算力的注意力机制。YOLOv11 是当前最先进的目标检测算法之一,在处理多目标、不同分辨率和复杂背景下表现优异。
-
定制化改造 :
修改目标:80 通用目标 objects -> 50 + 中药材 objects。
数据格式 YOLO 化:将中药材的XML 格式切换为 YOLO 格式。 -
基线模型性能 :
基线模型 YOLOv8,成功实现对 50 + 类中药材的高效检测,取得 81.1%的 mAP,顺利打通了基于 YOLO 的完整流程,包括数据预处理、模型训练与测试。
模型改进
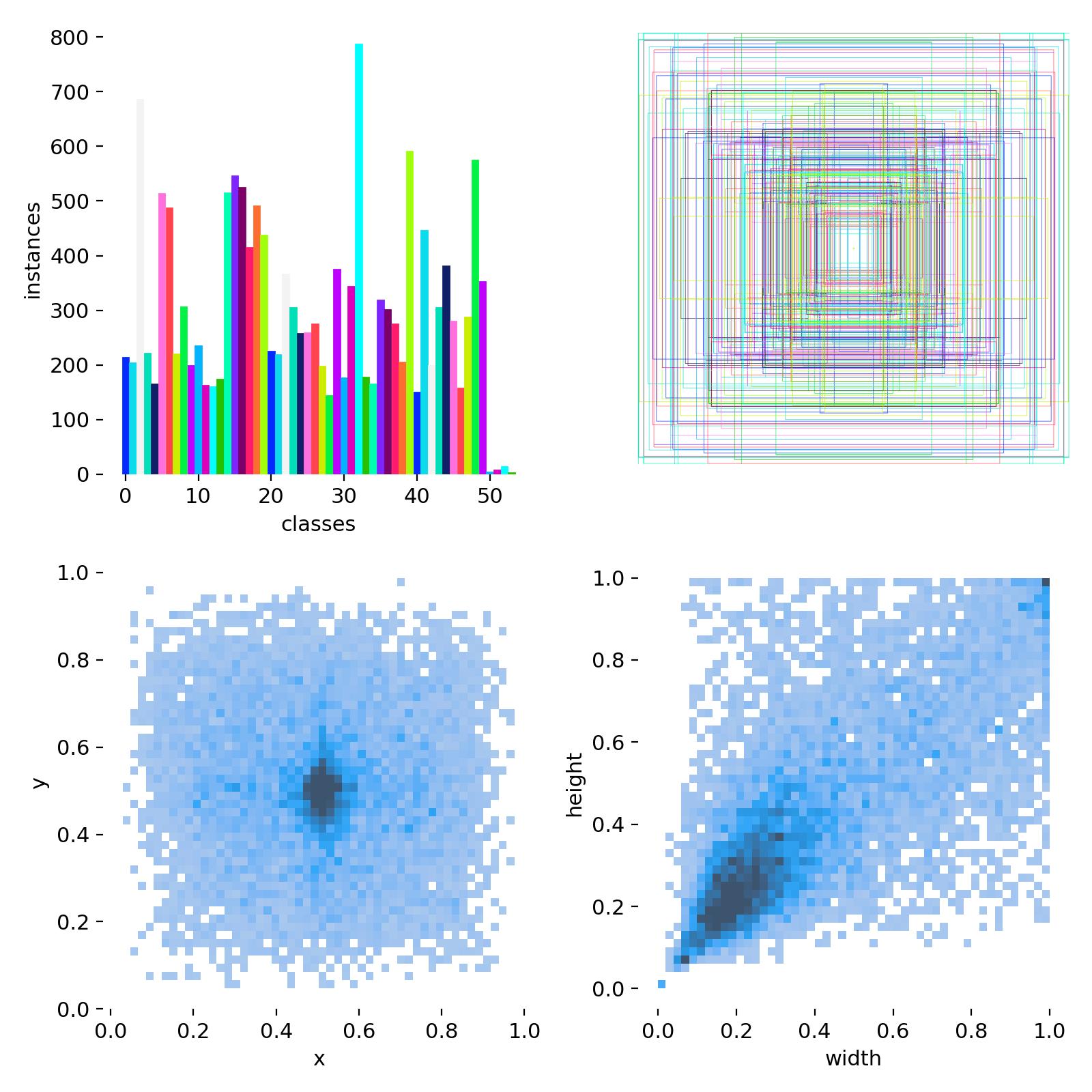



(一)baseline性能
Epoch GPU_mem box_loss cls_loss dfl_loss Instances Size120/120 6.91G 1.374 1.145 1.657 3 832: 100%|██████████| 482/482 [00:20<00:00, 23.48it/s]Class Images Instances Box(P R mAP50 mAP50-95): 100%|██████████| 32/32 [00:01<00:00, 19.14it/s]all 1005 2152 0.782 0.739 0.811 0.474120 epochs completed in 0.792 hours.
Optimizer stripped from runs/ChineseMedTrain/exp8/weights/last.pt, 5.5MB
Optimizer stripped from runs/ChineseMedTrain/exp8/weights/best.pt, 5.5MBValidating runs/ChineseMedTrain/exp8/weights/best.pt...
Ultralytics 8.3.7 🚀 Python-3.9.19 torch-2.0.1+cu117 CUDA:0 (NVIDIA GeForce RTX 4090, 24209MiB)
YOLO11n summary (fused): 238 layers, 2,591,902 parameters, 0 gradients, 6.4 GFLOPsClass Images Instances Box(P R mAP50 mAP50-95): 100%|██████████| 32/32 [00:02<00:00, 15.02it/s]all 1005 2152 0.788 0.735 0.811 0.474ginseng 34 57 0.869 0.772 0.864 0.483Leech 20 41 0.778 0.769 0.847 0.513JujubaeFructus 18 67 0.829 0.761 0.86 0.491LiliiBulbus 18 19 0.64 0.789 0.835 0.552CoptidisRhizoma 22 22 0.868 0.898 0.96 0.758MumeFructus 21 98 0.716 0.693 0.756 0.372MagnoliaBark 21 45 0.737 0.746 0.814 0.416Oyster 18 24 0.735 0.809 0.846 0.595Seahorse 14 33 0.835 0.424 0.493 0.274Luohanguo 17 21 0.834 0.714 0.793 0.593GlycyrrhizaUralensis 18 25 0.92 0.92 0.978 0.502Sanqi 32 42 0.753 0.714 0.761 0.544TetrapanacisMedulla 19 20 0.859 0.915 0.977 0.622CoicisSemen 24 35 0.88 0.628 0.823 0.492LyciiFructus 20 32 0.829 0.562 0.772 0.411TruestarAnise 18 60 0.853 0.679 0.894 0.376ClamShell 17 67 0.699 0.746 0.765 0.466Chuanxiong 28 69 0.782 0.623 0.766 0.372Garlic 24 70 0.801 0.748 0.793 0.341GinkgoBiloba 27 119 0.767 0.807 0.859 0.532ChrysanthemiFlos 13 20 0.786 0.7 0.734 0.436
AtractylodesMacrocephala 15 23 0.807 0.909 0.886 0.576JuglandisSemen 12 45 0.87 0.448 0.689 0.332TallGastrodiae 17 35 0.577 0.74 0.689 0.339TrionycisCarapax 15 22 0.666 0.636 0.749 0.515AngelicaRoot 18 35 0.78 0.886 0.89 0.538Hawthorn 21 47 0.683 0.366 0.565 0.253CrociStigma 20 22 0.951 0.874 0.948 0.523SerpentisPeriostracum 16 16 0.864 0.875 0.929 0.598EucommiaBark 17 32 0.844 0.781 0.841 0.484ImperataeRhizoma 21 22 0.904 0.909 0.944 0.579LoniceraJaponica 12 25 0.525 0.531 0.549 0.279Zhizi 20 128 0.806 0.336 0.589 0.242Scorpion 13 21 0.812 0.81 0.867 0.619HouttuyniaeHerba 16 16 0.952 1 0.995 0.596EupolyphagaSinensis 19 48 0.641 0.875 0.856 0.509OroxylumIndicum 31 67 0.827 0.821 0.886 0.458CurcumaLonga 34 63 0.718 0.726 0.738 0.444NelumbinisPlumula 17 20 0.797 0.7 0.748 0.458ArecaeSemen 22 66 0.668 0.424 0.71 0.352Scolopendra 19 25 0.801 0.6 0.667 0.437MoriFructus 22 64 0.725 0.688 0.687 0.3
FritillariaeCirrhosaeBulbus 24 26 0.747 0.846 0.87 0.561DioscoreaeRhizoma 23 34 0.896 0.757 0.911 0.45CicadaePeriostracum 17 41 0.824 0.927 0.914 0.531PiperCubeba 21 28 0.825 0.821 0.873 0.504BupleuriRadix 22 25 0.814 0.72 0.889 0.499AntelopeHom 18 48 0.771 0.839 0.853 0.556Pangdahai 19 71 0.859 0.769 0.882 0.575NelumbinisSemen 19 51 0.674 0.73 0.764 0.447
Speed: 0.2ms preprocess, 0.3ms inference, 0.0ms loss, 0.3ms postprocess per image
Results saved to runs/ChineseMedTrain/exp8
(二)baseline+ DETR head
提醒:在yolo11之后添加RT-DETR会失败;正确的思路是利用RT-DETR作为baseline
经过测试,采用RT-DETR检测头,导致训练速度降低4倍。
(三)baseline+ RepC3K2
改进的点:C3K2重参数化 Rep技术;

快速实现的 CSP 瓶颈模块(Faster Implementation of CSP Bottleneck)
基于 C2f 进一步优化,用于特征提取,支持可配置的卷积块(C3k 或普通 Bottleneck核心是通过模块化堆叠,平衡计算量与特征表达能力,常用于 YOLO 系列骨干网络
- Attributes:
m: 模块列表,根据 c3k 标志决定是用 C3k 还是 Bottleneck 构建内部结构 - Methods:
init: 初始化,配置输入输出通道、模块数量、是否用 C3k 等参数
forward: 前向传播,通过卷积、残差等操作提取并融合特征
Epoch GPU_mem box_loss cls_loss dfl_loss Instances Size120/120 13.9G 1.225 0.874 1.51 45 352: 100%|██████████| 241/241 [00:18<00:00, 12.76it/s]Class Images Instances Box(P R mAP50 mAP50-95): 100%|██████████| 16/16 [00:01<00:00, 9.69it/s]all 1005 2152 0.833 0.792 0.854 0.527120 epochs completed in 0.716 hours.
Optimizer stripped from runs/ChineseMedTrain/exp2/weights/last.pt, 5.6MB
Optimizer stripped from runs/ChineseMedTrain/exp2/weights/best.pt, 5.6MBValidating runs/ChineseMedTrain/exp2/weights/best.pt...
WARNING ⚠️ validating an untrained model YAML will result in 0 mAP.
Ultralytics 8.3.7 🚀 Python-3.9.19 torch-2.0.1+cu117 CUDA:0 (NVIDIA GeForce RTX 4090, 24209MiB)
YOLO11RepC3K2 summary (fused): 239 layers, 2,591,902 parameters, 0 gradients, 6.4 GFLOPsClass Images Instances Box(P R mAP50 mAP50-95): 100%|██████████| 16/16 [00:02<00:00, 6.57it/s]all 1005 2152 0.833 0.79 0.854 0.527ginseng 34 57 0.859 0.853 0.914 0.581Leech 20 41 0.759 0.845 0.884 0.621JujubaeFructus 18 67 0.891 0.856 0.911 0.545LiliiBulbus 18 19 0.82 0.895 0.898 0.563CoptidisRhizoma 22 22 0.868 0.895 0.97 0.799MumeFructus 21 98 0.747 0.694 0.784 0.408MagnoliaBark 21 45 0.889 0.844 0.906 0.521Oyster 18 24 0.756 0.917 0.929 0.685Seahorse 14 33 0.942 0.489 0.593 0.345Luohanguo 17 21 0.767 0.762 0.777 0.618GlycyrrhizaUralensis 18 25 0.886 1 0.989 0.505Sanqi 32 42 0.735 0.714 0.788 0.586TetrapanacisMedulla 19 20 0.959 0.95 0.993 0.626CoicisSemen 24 35 0.936 0.835 0.922 0.59LyciiFructus 20 32 0.832 0.562 0.715 0.428TruestarAnise 18 60 0.936 0.732 0.939 0.415ClamShell 17 67 0.806 0.672 0.801 0.493Chuanxiong 28 69 0.797 0.783 0.803 0.439Garlic 24 70 0.806 0.643 0.817 0.421GinkgoBiloba 27 119 0.86 0.823 0.899 0.572ChrysanthemiFlos 13 20 0.788 0.75 0.71 0.429
AtractylodesMacrocephala 15 23 0.858 0.826 0.902 0.603JuglandisSemen 12 45 0.906 0.639 0.83 0.395TallGastrodiae 17 35 0.726 0.758 0.799 0.436TrionycisCarapax 15 22 0.782 0.818 0.811 0.601AngelicaRoot 18 35 0.877 0.914 0.909 0.586Hawthorn 21 47 0.873 0.638 0.754 0.374CrociStigma 20 22 1 0.952 0.957 0.541SerpentisPeriostracum 16 16 0.853 0.938 0.966 0.736EucommiaBark 17 32 0.854 0.875 0.913 0.572ImperataeRhizoma 21 22 0.913 0.955 0.95 0.621LoniceraJaponica 12 25 0.547 0.6 0.695 0.331Zhizi 20 128 0.82 0.5 0.685 0.309Scorpion 13 21 0.833 0.857 0.873 0.606HouttuyniaeHerba 16 16 0.951 1 0.995 0.65EupolyphagaSinensis 19 48 0.76 0.958 0.88 0.561OroxylumIndicum 31 67 0.838 0.821 0.932 0.51CurcumaLonga 34 63 0.667 0.698 0.815 0.501NelumbinisPlumula 17 20 0.886 0.7 0.777 0.51ArecaeSemen 22 66 0.879 0.667 0.894 0.455Scolopendra 19 25 0.776 0.64 0.638 0.467MoriFructus 22 64 0.719 0.679 0.677 0.307
FritillariaeCirrhosaeBulbus 24 26 0.736 0.846 0.906 0.641DioscoreaeRhizoma 23 34 0.828 0.847 0.909 0.493CicadaePeriostracum 17 41 0.828 0.937 0.914 0.581PiperCubeba 21 28 0.92 0.821 0.869 0.576BupleuriRadix 22 25 0.938 0.8 0.924 0.507AntelopeHom 18 48 0.817 0.812 0.911 0.584Pangdahai 19 71 0.869 0.746 0.874 0.583NelumbinisSemen 19 51 0.763 0.759 0.803 0.512
Speed: 0.4ms preprocess, 0.4ms inference, 0.0ms loss, 0.3ms postprocess per image
Results saved to runs/ChineseMedTrain/exp2
(四)baseline+ RepC3K2 + SimSPPF
改进的点:SimSPPF简化SPPF模块;

简化版空间金字塔池化模块(Simplified SPP
对输入特征进行多尺度池化与融合,提升模型对不同尺度目标的感知能力常用于 YOLO 等检测模型 Neck 部分,增强特征鲁棒性
- Attributes:
sppf: 实际执行池化、卷积操作的子模块(SPPFModule) - Methods:
init: 初始化,配置输入输出通道、池化核大小等
forward: 直接调用 sppf 子模块完成前向传播
engine/trainer: task=detect, mode=train, model=yolo11RepC3K2SimSPPF.yaml, data=ultralytics/cfg/datasets/originalChineseMed50.yaml, epochs=120, time=None, patience=150, batch=32, imgsz=640, save=True, save_period=10, cache=False, device=0, workers=8, project=runs/ChineseMedTrain, name=exp3, exist_ok=False, pretrained=/home/wqt/Projects/yolov11/ultralytics/runs/ChineseMedTrain/exp2/weights/best.pt, optimizer=auto, verbose=True, seed=0, deterministic=True, single_cls=False, rect=False, cos_lr=True, close_mosaic=10, resume=False, amp=True, fraction=1.0, profile=False, freeze=None, multi_scale=True, overlap_mask=True, mask_ratio=4, dropout=0.0, val=True, split=val, save_json=False, save_hybrid=False, conf=None, iou=0.7, max_det=300, half=False, dnn=False, plots=True, source=None, vid_stride=1, stream_buffer=False, visualize=False, augment=False, agnostic_nms=False, classes=None, retina_masks=False, embed=None, show=False, save_frames=False, save_txt=False, save_conf=False, save_crop=False, show_labels=True, show_conf=True, show_boxes=True, line_width=None, format=torchscript, keras=False, optimize=False, int8=False, dynamic=False, simplify=True, opset=None, workspace=4, nms=False, lr0=0.01, lrf=0.01, momentum=0.937, weight_decay=0.0005, warmup_epochs=3.0, warmup_momentum=0.8, warmup_bias_lr=0.1, box=7.5, cls=0.5, dfl=1.5, pose=12.0, kobj=1.0, label_smoothing=0.0, nbs=64, hsv_h=0.015, hsv_s=0.7, hsv_v=0.4, degrees=0.0, translate=0.1, scale=0.9, shear=0.0, perspective=0.0, flipud=0.0, fliplr=0.5, bgr=0.0, mosaic=1.0, mixup=0.2, copy_paste=0.0, copy_paste_mode=flip, auto_augment=randaugment, erasing=0.4, crop_fraction=1.0, cfg=None, tracker=botsort.yaml, save_dir=runs/ChineseMedTrain/exp3
Overriding model.yaml nc=80 with nc=50
WARNING ⚠️ no model scale passed. Assuming scale='n'.from n params module arguments 0 -1 1 464 ultralytics.nn.modules.conv.Conv [3, 16, 3, 2] 1 -1 1 4672 ultralytics.nn.modules.conv.Conv [16, 32, 3, 2] 2 -1 1 6640 ultralytics.nn.modules.block.RepC3k2 [32, 64, 1, False, 0.25] 3 -1 1 36992 ultralytics.nn.modules.conv.Conv [64, 64, 3, 2] 4 -1 1 26080 ultralytics.nn.modules.block.RepC3k2 [64, 128, 1, False, 0.25] 5 -1 1 147712 ultralytics.nn.modules.conv.Conv [128, 128, 3, 2] 6 -1 1 89216 ultralytics.nn.modules.block.RepC3k2 [128, 128, 1, True] 7 -1 1 295424 ultralytics.nn.modules.conv.Conv [128, 256, 3, 2] 8 -1 1 354560 ultralytics.nn.modules.block.RepC3k2 [256, 256, 1, True] 9 -1 1 164608 ultralytics.nn.modules.block.SimSPPF [256, 256, 5]
10 -1 1 249728 ultralytics.nn.modules.block.C2PSA [256, 256, 1]
11 -1 1 0 torch.nn.modules.upsampling.Upsample [None, 2, 'nearest']
12 [-1, 6] 1 0 ultralytics.nn.modules.conv.Concat [1]
13 -1 1 111296 ultralytics.nn.modules.block.RepC3k2 [384, 128, 1, False]
14 -1 1 0 torch.nn.modules.upsampling.Upsample [None, 2, 'nearest']
15 [-1, 4] 1 0 ultralytics.nn.modules.conv.Concat [1]
16 -1 1 32096 ultralytics.nn.modules.block.RepC3k2 [256, 64, 1, False]
17 -1 1 36992 ultralytics.nn.modules.conv.Conv [64, 64, 3, 2]
18 [-1, 13] 1 0 ultralytics.nn.modules.conv.Concat [1]
19 -1 1 86720 ultralytics.nn.modules.block.RepC3k2 [192, 128, 1, False]
20 -1 1 147712 ultralytics.nn.modules.conv.Conv [128, 128, 3, 2]
21 [-1, 10] 1 0 ultralytics.nn.modules.conv.Concat [1]
22 -1 1 387328 ultralytics.nn.modules.block.RepC3k2 [384, 256, 1, True]
23 [16, 19, 22] 1 440422 ultralytics.nn.modules.head.Detect [50, [64, 128, 256]]
YOLO11RepC3K2SimSPPF summary: 360 layers, 2,618,662 parameters, 2,618,646 gradients, 6.5 GFLOPs
Epoch GPU_mem box_loss cls_loss dfl_loss Instances Size120/120 13.8G 1.149 0.7983 1.44 45 352: 100%|██████████| 241/241 [00:19<00:00, 12.35it/s]Class Images Instances Box(P R mAP50 mAP50-95): 100%|██████████| 16/16 [00:01<00:00, 9.26it/s]all 1005 2152 0.867 0.799 0.872 0.549120 epochs completed in 0.714 hours.
Optimizer stripped from runs/ChineseMedTrain/exp3/weights/last.pt, 5.6MB
Optimizer stripped from runs/ChineseMedTrain/exp3/weights/best.pt, 5.6MBValidating runs/ChineseMedTrain/exp3/weights/best.pt...
WARNING ⚠️ validating an untrained model YAML will result in 0 mAP.
Ultralytics 8.3.7 🚀 Python-3.9.19 torch-2.0.1+cu117 CUDA:0 (NVIDIA GeForce RTX 4090, 24209MiB)
YOLO11RepC3K2SimSPPF summary (fused): 245 layers, 2,592,286 parameters, 0 gradients, 6.4 GFLOPsClass Images Instances Box(P R mAP50 mAP50-95): 100%|██████████| 16/16 [00:02<00:00, 6.30it/s]all 1005 2152 0.867 0.802 0.871 0.549ginseng 34 57 0.888 0.833 0.907 0.587Leech 20 41 0.815 0.902 0.932 0.65JujubaeFructus 18 67 0.904 0.839 0.968 0.585LiliiBulbus 18 19 0.946 0.895 0.9 0.588CoptidisRhizoma 22 22 0.874 0.955 0.984 0.824MumeFructus 21 98 0.746 0.781 0.802 0.411MagnoliaBark 21 45 0.848 0.865 0.932 0.576Oyster 18 24 0.841 1 0.971 0.706Seahorse 14 33 0.835 0.613 0.627 0.37Luohanguo 17 21 0.876 0.667 0.79 0.613GlycyrrhizaUralensis 18 25 0.926 0.995 0.984 0.537Sanqi 32 42 0.908 0.643 0.818 0.625TetrapanacisMedulla 19 20 0.984 0.95 0.987 0.672CoicisSemen 24 35 0.823 0.829 0.907 0.6LyciiFructus 20 32 0.865 0.599 0.758 0.459TruestarAnise 18 60 1 0.678 0.953 0.432ClamShell 17 67 0.806 0.731 0.841 0.538Chuanxiong 28 69 0.775 0.768 0.796 0.427Garlic 24 70 0.848 0.715 0.858 0.435GinkgoBiloba 27 119 0.847 0.866 0.914 0.61ChrysanthemiFlos 13 20 0.937 0.745 0.788 0.488
AtractylodesMacrocephala 15 23 0.826 0.828 0.893 0.614JuglandisSemen 12 45 0.912 0.694 0.785 0.389TallGastrodiae 17 35 0.75 0.743 0.764 0.408TrionycisCarapax 15 22 0.866 0.818 0.865 0.61AngelicaRoot 18 35 0.894 0.914 0.904 0.582Hawthorn 21 47 0.979 0.66 0.766 0.434CrociStigma 20 22 0.91 0.864 0.934 0.529SerpentisPeriostracum 16 16 0.974 0.938 0.988 0.727EucommiaBark 17 32 0.942 0.812 0.942 0.629ImperataeRhizoma 21 22 0.879 0.909 0.974 0.653LoniceraJaponica 12 25 0.779 0.565 0.707 0.383Zhizi 20 128 0.923 0.563 0.765 0.355Scorpion 13 21 0.845 0.905 0.935 0.679HouttuyniaeHerba 16 16 0.846 0.938 0.986 0.662EupolyphagaSinensis 19 48 0.757 0.976 0.924 0.62OroxylumIndicum 31 67 0.82 0.885 0.872 0.474CurcumaLonga 34 63 0.836 0.73 0.857 0.521NelumbinisPlumula 17 20 0.708 0.7 0.778 0.514ArecaeSemen 22 66 0.942 0.745 0.923 0.471Scolopendra 19 25 0.878 0.64 0.669 0.471MoriFructus 22 64 0.77 0.719 0.758 0.355
FritillariaeCirrhosaeBulbus 24 26 0.841 0.885 0.936 0.65DioscoreaeRhizoma 23 34 0.846 0.811 0.919 0.528CicadaePeriostracum 17 41 0.865 0.951 0.931 0.636PiperCubeba 21 28 0.865 0.857 0.91 0.579BupleuriRadix 22 25 1 0.739 0.907 0.557AntelopeHom 18 48 0.929 0.771 0.899 0.623Pangdahai 19 71 0.872 0.861 0.897 0.603NelumbinisSemen 19 51 0.787 0.804 0.761 0.473
Speed: 0.3ms preprocess, 0.4ms inference, 0.0ms loss, 0.3ms postprocess per image
Results saved to runs/ChineseMedTrain/exp3
(五)baseline+ RepC3K2 + SimSPPF + LK-C2PSA
改进的点:将PSA模块中的Attention修改为Deformable-LK Attention,即可变形的大核Attention;

位置敏感注意力模块(Position - Sensitive Attention block)
用于神经网络特征提取整合多头注意力(Multi - head attention)与前馈神经网络(Feed - forward),支持残差连接(shortcut )
- Attributes:
1. attn: 多头注意力模块,实现注意力机制捕捉特征关联
2. ffn: 前馈神经网络模块,对注意力输出进一步特征变换
3. add: 是否添加残差连接的标志,残差有助于缓解梯度消失、增强特征复用 - Methods:
1. init: 初始化模块,配置注意力、前馈网络及残差连接
2. forward: 前向传播,先过注意力再经过前馈网络,带残差时做 shortcut 相加 - Example:
psa_block = LKPSA(c=128, attn_ratio=0.5, num_heads=4, shortcut=True)
input_tensor = torch.randn(1, 128, 32, 32)
output = psa_block(input_tensor)
Epoch GPU_mem box_loss cls_loss dfl_loss Instances Size120/120 12.8G 1.122 0.7926 1.414 45 352: 100%|██████████| 241/241 [00:41<00:00, 5.74it/s]Class Images Instances Box(P R mAP50 mAP50-95): 100%|██████████| 16/16 [00:03<00:00, 4.89it/s]all 1005 2152 0.865 0.805 0.867 0.556120 epochs completed in 1.539 hours.
Optimizer stripped from runs/ChineseMedTrain/exp4/weights/last.pt, 7.0MB
Optimizer stripped from runs/ChineseMedTrain/exp4/weights/best.pt, 7.0MBValidating runs/ChineseMedTrain/exp4/weights/best.pt...
WARNING ⚠️ validating an untrained model YAML will result in 0 mAP.
Ultralytics 8.3.7 🚀 Python-3.9.19 torch-2.0.1+cu117 CUDA:0 (NVIDIA GeForce RTX 4090, 24209MiB)
YOLO11RepC3K2SimSPPF_LKPSA summary (fused): 243 layers, 3,342,258 parameters, 0 gradients, 7.0 GFLOPsClass Images Instances Box(P R mAP50 mAP50-95): 100%|██████████| 16/16 [00:04<00:00, 3.94it/s]all 1005 2152 0.859 0.806 0.867 0.557ginseng 34 57 0.915 0.76 0.912 0.592Leech 20 41 0.825 0.927 0.926 0.637JujubaeFructus 18 67 0.875 0.91 0.932 0.572LiliiBulbus 18 19 0.917 0.842 0.906 0.61CoptidisRhizoma 22 22 0.912 1 0.992 0.839MumeFructus 21 98 0.79 0.816 0.826 0.46MagnoliaBark 21 45 0.866 0.933 0.961 0.587Oyster 18 24 0.74 0.958 0.947 0.713Seahorse 14 33 0.928 0.485 0.612 0.376Luohanguo 17 21 0.785 0.667 0.816 0.611GlycyrrhizaUralensis 18 25 0.924 0.98 0.973 0.547Sanqi 32 42 0.946 0.714 0.867 0.669TetrapanacisMedulla 19 20 1 0.968 0.995 0.684CoicisSemen 24 35 0.881 0.914 0.951 0.604LyciiFructus 20 32 0.835 0.625 0.783 0.463TruestarAnise 18 60 0.873 0.783 0.897 0.406ClamShell 17 67 0.758 0.642 0.745 0.489Chuanxiong 28 69 0.795 0.754 0.796 0.439Garlic 24 70 0.931 0.767 0.89 0.472GinkgoBiloba 27 119 0.814 0.866 0.917 0.584ChrysanthemiFlos 13 20 0.833 0.8 0.798 0.495
AtractylodesMacrocephala 15 23 0.742 0.826 0.813 0.595JuglandisSemen 12 45 0.899 0.592 0.767 0.391TallGastrodiae 17 35 0.843 0.686 0.818 0.45TrionycisCarapax 15 22 0.856 0.818 0.877 0.682AngelicaRoot 18 35 0.893 0.914 0.898 0.622Hawthorn 21 47 0.904 0.681 0.781 0.466CrociStigma 20 22 0.817 0.909 0.94 0.529SerpentisPeriostracum 16 16 0.886 0.973 0.97 0.751EucommiaBark 17 32 0.854 0.906 0.92 0.597ImperataeRhizoma 21 22 0.936 0.955 0.954 0.662LoniceraJaponica 12 25 0.604 0.64 0.642 0.315Zhizi 20 128 0.879 0.509 0.711 0.339Scorpion 13 21 0.861 0.905 0.967 0.706HouttuyniaeHerba 16 16 0.975 0.938 0.972 0.706EupolyphagaSinensis 19 48 0.76 1 0.947 0.614OroxylumIndicum 31 67 0.861 0.776 0.822 0.484CurcumaLonga 34 63 0.793 0.762 0.856 0.533NelumbinisPlumula 17 20 0.801 0.7 0.727 0.517ArecaeSemen 22 66 0.937 0.667 0.901 0.475Scolopendra 19 25 0.913 0.64 0.641 0.492MoriFructus 22 64 0.832 0.695 0.786 0.388
FritillariaeCirrhosaeBulbus 24 26 0.898 0.885 0.932 0.658DioscoreaeRhizoma 23 34 0.904 0.827 0.968 0.522CicadaePeriostracum 17 41 0.865 0.938 0.946 0.66PiperCubeba 21 28 0.76 0.791 0.905 0.583BupleuriRadix 22 25 0.955 0.854 0.929 0.576AntelopeHom 18 48 0.979 0.812 0.935 0.663Pangdahai 19 71 0.791 0.789 0.844 0.569NelumbinisSemen 19 51 0.787 0.796 0.734 0.435
Speed: 0.3ms preprocess, 2.0ms inference, 0.0ms loss, 0.2ms postprocess per image
Results saved to runs/ChineseMedTrain/exp4
该改进的亮点是,mAP50-95的性能得到提升,说明它在复杂场景下的识别率有所提升。
(六)baseline+ RepC3K2 + SimSPPF + LK-C2PSA TriAttentionPSA
改进的点:将PSA模块中的Attention修改为 采用以下博客中的技术,即TriAttention,三重注意力机制来改进PSA模块;

基于三元组注意力(Triplet Attention)的位置敏感注意力模块
同样包含多头注意力(这里是 TripletAttention 实现)、前馈网络及残差连接逻辑
功能与 LKPSA 类似,核心差异在注意力机制的具体计算方式(三元组注意力侧重多维度关联)
Attributes、Methods 逻辑同 LKPSA,仅注意力实例化为 TripletAttention
0 -1 1 464 ultralytics.nn.modules.conv.Conv [3, 16, 3, 2] 1 -1 1 4672 ultralytics.nn.modules.conv.Conv [16, 32, 3, 2] 2 -1 1 6640 ultralytics.nn.modules.block.RepC3k2 [32, 64, 1, False, 0.25] 3 -1 1 36992 ultralytics.nn.modules.conv.Conv [64, 64, 3, 2] 4 -1 1 26080 ultralytics.nn.modules.block.RepC3k2 [64, 128, 1, False, 0.25] 5 -1 1 147712 ultralytics.nn.modules.conv.Conv [128, 128, 3, 2] 6 -1 1 89216 ultralytics.nn.modules.block.RepC3k2 [128, 128, 1, True] 7 -1 1 295424 ultralytics.nn.modules.conv.Conv [128, 256, 3, 2] 8 -1 1 354560 ultralytics.nn.modules.block.RepC3k2 [256, 256, 1, True] 9 -1 1 164608 ultralytics.nn.modules.block.SimSPPF [256, 256, 5] 10 -1 1 198700 ultralytics.nn.modules.block.TriC2PSA [256, 256, 1] 11 -1 1 0 torch.nn.modules.upsampling.Upsample [None, 2, 'nearest'] 12 [-1, 6] 1 0 ultralytics.nn.modules.conv.Concat [1] 13 -1 1 111296 ultralytics.nn.modules.block.RepC3k2 [384, 128, 1, False] 14 -1 1 0 torch.nn.modules.upsampling.Upsample [None, 2, 'nearest'] 15 [-1, 4] 1 0 ultralytics.nn.modules.conv.Concat [1] 16 -1 1 32096 ultralytics.nn.modules.block.RepC3k2 [256, 64, 1, False] 17 -1 1 36992 ultralytics.nn.modules.conv.Conv [64, 64, 3, 2] 18 [-1, 13] 1 0 ultralytics.nn.modules.conv.Concat [1] 19 -1 1 86720 ultralytics.nn.modules.block.RepC3k2 [192, 128, 1, False] 20 -1 1 147712 ultralytics.nn.modules.conv.Conv [128, 128, 3, 2] 21 [-1, 10] 1 0 ultralytics.nn.modules.conv.Concat [1] 22 -1 1 387328 ultralytics.nn.modules.block.RepC3k2 [384, 256, 1, True] 23 [16, 19, 22] 1 440422 ultralytics.nn.modules.head.Detect [50, [64, 128, 256]]
YOLO11RepC3K2SimSPPF_TriPSA summary: 363 layers, 2,567,634 parameters, 2,567,618 gradients, 6.5 GFLOPsTransferred 484/535 items from pretrained weights
以下是运行的结果:
Epoch GPU_mem box_loss cls_loss dfl_loss Instances Size120/120 13.5G 1.127 0.8044 1.416 45 352: 100%|██████████| 241/241 [00:18<00:00, 12.77it/s]Class Images Instances Box(P R mAP50 mAP50-95): 100%|██████████| 16/16 [00:01<00:00, 9.72it/s]all 1005 2152 0.87 0.8 0.865 0.556120 epochs completed in 0.713 hours.
Optimizer stripped from runs/ChineseMedTrain/exp5/weights/last.pt, 5.5MB
Optimizer stripped from runs/ChineseMedTrain/exp5/weights/best.pt, 5.5MBValidating runs/ChineseMedTrain/exp5/weights/best.pt...
WARNING ⚠️ validating an untrained model YAML will result in 0 mAP.
Ultralytics 8.3.7 🚀 Python-3.9.19 torch-2.0.1+cu117 CUDA:0 (NVIDIA GeForce RTX 4090, 24209MiB)
YOLO11RepC3K2SimSPPF_TriPSA summary (fused): 251 layers, 2,541,770 parameters, 0 gradients, 6.3 GFLOPsClass Images Instances Box(P R mAP50 mAP50-95): 100%|██████████| 16/16 [00:02<00:00, 6.52it/s]all 1005 2152 0.87 0.797 0.864 0.557ginseng 34 57 0.894 0.739 0.911 0.616Leech 20 41 0.881 0.902 0.931 0.66JujubaeFructus 18 67 0.903 0.834 0.921 0.579LiliiBulbus 18 19 0.889 0.841 0.873 0.578CoptidisRhizoma 22 22 0.911 0.926 0.983 0.822MumeFructus 21 98 0.76 0.786 0.817 0.427MagnoliaBark 21 45 0.809 0.844 0.92 0.572Oyster 18 24 0.83 0.917 0.942 0.697Seahorse 14 33 0.941 0.486 0.601 0.387Luohanguo 17 21 0.89 0.768 0.851 0.636GlycyrrhizaUralensis 18 25 0.926 0.998 0.978 0.594Sanqi 32 42 0.902 0.643 0.82 0.652TetrapanacisMedulla 19 20 0.981 0.95 0.99 0.678CoicisSemen 24 35 0.884 0.857 0.924 0.617LyciiFructus 20 32 0.868 0.617 0.785 0.47TruestarAnise 18 60 0.961 0.827 0.94 0.445ClamShell 17 67 0.783 0.701 0.765 0.495Chuanxiong 28 69 0.832 0.754 0.803 0.434Garlic 24 70 0.824 0.686 0.816 0.453GinkgoBiloba 27 119 0.841 0.888 0.922 0.608ChrysanthemiFlos 13 20 0.84 0.789 0.782 0.511
AtractylodesMacrocephala 15 23 0.782 0.826 0.843 0.583JuglandisSemen 12 45 0.922 0.6 0.711 0.406TallGastrodiae 17 35 0.787 0.629 0.759 0.409TrionycisCarapax 15 22 0.82 0.831 0.865 0.652AngelicaRoot 18 35 0.894 0.914 0.902 0.606Hawthorn 21 47 0.896 0.681 0.761 0.458CrociStigma 20 22 0.899 0.909 0.931 0.529
SerpentisPeriostracum 16 16 0.938 0.939 0.988 0.754EucommiaBark 17 32 0.939 0.938 0.948 0.619ImperataeRhizoma 21 22 0.942 0.955 0.989 0.654LoniceraJaponica 12 25 0.751 0.64 0.72 0.379Zhizi 20 128 0.827 0.522 0.709 0.336Scorpion 13 21 0.883 0.952 0.955 0.678HouttuyniaeHerba 16 16 0.982 0.938 0.981 0.662EupolyphagaSinensis 19 48 0.716 0.979 0.917 0.635OroxylumIndicum 31 67 0.872 0.761 0.82 0.487CurcumaLonga 34 63 0.801 0.683 0.856 0.553NelumbinisPlumula 17 20 0.794 0.7 0.74 0.504ArecaeSemen 22 66 0.953 0.697 0.926 0.494Scolopendra 19 25 0.898 0.6 0.685 0.518MoriFructus 22 64 0.861 0.675 0.8 0.382
FritillariaeCirrhosaeBulbus 24 26 0.836 0.885 0.902 0.641DioscoreaeRhizoma 23 34 0.842 0.824 0.936 0.55CicadaePeriostracum 17 41 0.857 0.951 0.893 0.6PiperCubeba 21 28 0.886 0.832 0.908 0.582BupleuriRadix 22 25 0.97 0.88 0.92 0.555AntelopeHom 18 48 0.997 0.833 0.93 0.659Pangdahai 19 71 0.812 0.789 0.844 0.574NelumbinisSemen 19 51 0.807 0.74 0.77 0.457
Speed: 0.4ms preprocess, 0.3ms inference, 0.0ms loss, 0.2ms postprocess per image
Results saved to runs/ChineseMedTrain/exp5
(七)baseline+ RepC3K2 + SimSPPF + LK-C2PSA TriAttentionPSA+ newLabels
改进的点:将PSA模块中的Attention修改为 采用以下博客中的技术,即TriAttention,三重注意力机制来改进PSA模块;
https://blog.csdn.net/m0_63774211/article/details/145569867
Epoch GPU_mem box_loss cls_loss dfl_loss Instances Size150/150 13.5G 0.9872 0.6845 1.308 56 864: 100%|██████████| 241/241 [00:20<00:00, 11.98it/s]Class Images Instances Box(P R mAP50 mAP50-95): 100%|██████████| 16/16 [00:01<00:00, 9.39it/s]all 1015 2299 0.869 0.751 0.813 0.548150 epochs completed in 0.894 hours.
Optimizer stripped from runs/ChineseMedTrain/exp8/weights/last.pt, 5.5MB
Optimizer stripped from runs/ChineseMedTrain/exp8/weights/best.pt, 5.5MBValidating runs/ChineseMedTrain/exp8/weights/best.pt...
Ultralytics 8.3.7 🚀 Python-3.9.19 torch-2.0.1+cu117 CUDA:0 (NVIDIA GeForce RTX 4090, 24209MiB)
YOLO11RepC3K2SimSPPF_TriPSA summary (fused): 251 layers, 2,542,745 parameters, 0 gradients, 6.4 GFLOPsClass Images Instances Box(P R mAP50 mAP50-95): 100%|██████████| 16/16 [00:02<00:00, 5.81it/s]all 1015 2299 0.865 0.754 0.812 0.546ginseng 35 58 0.92 0.741 0.899 0.643Leech 20 41 0.935 0.902 0.953 0.706JujubaeFructus 21 82 0.769 0.78 0.87 0.537LiliiBulbus 19 26 0.531 0.808 0.78 0.525CoptidisRhizoma 22 22 0.989 1 0.995 0.816MumeFructus 21 98 0.767 0.805 0.817 0.476MagnoliaBark 21 45 0.861 0.911 0.951 0.641Oyster 19 34 0.833 0.647 0.796 0.562Seahorse 14 33 0.934 0.545 0.594 0.384Luohanguo 17 21 0.943 0.796 0.829 0.672GlycyrrhizaUralensis 18 25 0.967 1 0.995 0.688Sanqi 32 42 0.935 0.643 0.824 0.651TetrapanacisMedulla 19 20 0.987 1 0.995 0.717CoicisSemen 24 35 0.901 0.857 0.95 0.676LyciiFructus 23 60 0.717 0.467 0.569 0.344TruestarAnise 18 60 0.958 0.754 0.964 0.497ClamShell 17 67 0.919 0.687 0.853 0.557Chuanxiong 28 69 0.828 0.797 0.849 0.549Garlic 24 70 0.91 0.722 0.887 0.494GinkgoBiloba 27 119 0.872 0.857 0.912 0.608ChrysanthemiFlos 13 20 0.893 0.7 0.786 0.539
AtractylodesMacrocephala 15 23 0.885 0.826 0.854 0.631JuglandisSemen 12 45 0.922 0.667 0.795 0.456TallGastrodiae 18 44 0.723 0.594 0.705 0.411TrionycisCarapax 16 32 0.704 0.596 0.698 0.507AngelicaRoot 18 35 0.855 0.914 0.914 0.663Hawthorn 21 47 0.921 0.744 0.796 0.505CrociStigma 20 22 0.867 0.89 0.929 0.601SerpentisPeriostracum 16 16 0.937 0.929 0.952 0.745EucommiaBark 17 32 0.901 0.938 0.942 0.664ImperataeRhizoma 21 22 0.948 1 0.989 0.712LoniceraJaponica 12 25 0.762 0.64 0.677 0.363Zhizi 21 132 0.82 0.623 0.753 0.426Scorpion 13 21 0.9 0.905 0.906 0.729HouttuyniaeHerba 16 16 0.952 0.938 0.991 0.747EupolyphagaSinensis 19 48 0.909 0.958 0.941 0.655OroxylumIndicum 31 67 0.929 0.779 0.866 0.58CurcumaLonga 34 63 0.893 0.746 0.841 0.576NelumbinisPlumula 17 20 0.888 0.85 0.874 0.603ArecaeSemen 22 66 0.942 0.733 0.924 0.558Scolopendra 19 25 0.888 0.634 0.642 0.519MoriFructus 23 74 0.844 0.66 0.745 0.421
FritillariaeCirrhosaeBulbus 26 42 0.586 0.844 0.795 0.522DioscoreaeRhizoma 23 34 0.938 0.896 0.965 0.544CicadaePeriostracum 17 41 0.826 0.951 0.941 0.67PiperCubeba 21 28 0.864 0.908 0.927 0.603BupleuriRadix 22 25 0.917 0.88 0.924 0.56AntelopeHom 18 48 0.925 0.688 0.881 0.615Pangdahai 19 71 0.867 0.845 0.869 0.608NelumbinisSemen 20 61 0.672 0.704 0.682 0.469Terminaliae Fructus 1 4 1 0 0.404 0.219
Aurantii Fructus Immaturus 1 7 0.373 1 0.601 0.314Poria 1 14 1 0 0.0514 0.0197
Processed Polygoni Multiflori Radix 1 2 1 0 0 0
Speed: 0.3ms preprocess, 0.4ms inference, 0.0ms loss, 0.4ms postprocess per image
Results saved to runs/ChineseMedTrain/exp8
四、应用场景与价值
应用场景
- 中药材科研与教育
- 辅助科研机构对珍稀药材标本进行自动化标注与特征提取,构建智能检索数据库。
- 为中医药院校提供交互式识别系统,帮助学生快速掌握。
- 应用价值
- 经济价值:该系统可显著降低中药材生产与加工企业的人力成本,提高分拣和检测效率,从而增强企业盈利能力。同时,精准的检测有助于提升产品质量,杜绝造假现象,进而增强企业信誉与竞争力,推动贸易增长。
- 社会价值:在中药材科研与教育领域,灵识本草能够为科研机构和院校提供高效、准确的辅助工具,加快科研进程,提高教学效果,为培养更多专业人才奠定基础。此外,通过严控中药材质量源头风险,守护用药安全,为人们的健康保驾护航,助推健康中国建设。还有助于发展中药材产业,健全农村产业体系,激发乡村振兴动能,带动农村经济发展和农民增收致富。
总结
灵识本草项目的成功实施,为中药材检测领域带来了创新性的解决方案。从项目的背景、目标,到实施过程、技术改进,再到最终的应用场景与价值,每一个环节都体现了团队对行业痛点的深入洞察和对技术创新的不懈追求。
在项目过程中,我们攻克了多项技术难题,通过深度学习技术的引入和优化,实现了中药材检测的高精度和快速度。这不仅提高了中药材行业的检测效率和质量,还为整个行业的数字化转型提供了有力支持。
未来,随着技术的不断发展和完善,灵识本草有望在更广泛的领域发挥其价值,为推动中药材产业的高质量发展,为保障人们的健康福祉,为促进乡村振兴战略的实施,持续贡献更多的力量。我们期待灵识本草能够在中药材行业的舞台上绽放出更加耀眼的光芒,书写属于中医药产业的新篇章。







